Autonomous nitrogen fixation of gramineous plants was realized by synthetic biology
Recently, The research group of Professor Chen Sanfeng of the College of Biology of China Agricultural University was published in the well-known academic journal "Trends in Biotechnology" (Trends in Biotechnology), which is owned by Cell Press The paper is published online in Biotechnology under the title "Using synthetic biology to express nitrogenase synthesis Pathways in Rice and Overcoming nitrogenase instability in plant cytoplasm.
biosynthesis pathway in rice and to overcome barriers of nitrogenase instability in plant cytosol) Research paper.
In this study, 13 nitrogen-fixing genes (about 40 kb) from 2 nitrogen-fixing bacteria were introduced into rice through multi-gene assembly, single transformation, cotransformation and hybridization.
Resequencing of the rice genome showed that the 13 nitrogen-fixing genes were integrated into 2 sites on chromosome 1 of rice. qRT-PCR and Western blot analysis showed that nitrogen-fixing genes could be transcribed and expressed in rice, and NifDK tetramer of nitrogen-fixing enzyme could be formed, and the problem of NifH protein being cut by endoprotein enzyme in plant cytoplasm was found and solved, which provided an important reference for realizing "autonomous nitrogen fixation" in grasses.

Nitrogen-fixing bacteria use the nitrogenase in the body to reduce N2 in the air to ammonia at normal temperature and pressure, which is called Biological nitrogen fixation.
Due to the lack of nodule symbiotic nitrogen fixation system, the high yield and stable yield of gramineous plants are highly dependent on chemical nitrogen fertilizer.
It is an effective way to reduce the amount of nitrogen fertilizer by introducing the nitrogenase synthesis pathway directly into gramineous plants to achieve "autonomous nitrogen fixation".
Nitrogenase is a very complex metalloenzyme composed of 2 protein components: ferritin (also known as NifH protein, encoded by the nifH gene) and molybdoferrin (also known as NifDK protein, encoded by the nifD and nifK genes).
In addition, three metal clusters are required for the synthesis of nitrogenase: [Fe4S4] cluster, P-cluster and FeMo-cofactor [MO-7 Fe-9S-C-Homocitrate].
FeMo-cofactor is located on the NifDK protein and is the site of N2 complexation and reduction, while the other two metal clusters function to transfer electrons.
The synthesis of nitrogenase requires at least 11 nitrogen-fixing genes [nifH, nifD, nifK, nifB, nifE, nifN, nifV, nifX, nifQ (or hesA), nifU, and nifS].
Heterologous expression of nitrogenase in plants faces many challenges: simultaneous introduction of multiple nitrogen fixing genes into plants, coordinated expression of multiple nitrogen fixing genes in plants, and stability of nitrogenase-related proteins in plants.
In this study, 13 genes related to nitrogenase synthesis were identified for the first time:
11 genes from Paenibacillus polymyxa (nifH, nifD, nifK, nifX, nifE, nifB, nifN, nifV, hesA, groES, and groEL) Two genes (nif U and nif S) from Klebsiella oxytoca were constructed in three plant expression vectors and introduced into rice through agrobacterium-mediated single and cotransformation.
The T1 generation transgenic rice was hybridized, and the first generation (F1) hybrid rice containing all 13 nitrogen-fixing genes was screened, and the fourth generation (F4) transgenic rice plants were obtained through continuous self-cross of rice.
Through PCR and RT-PCR analysis, 55 transgenic rice lines carrying all 13 genes related to nitrogenase synthesis were identified from 256 F4 generation transgenic rice lines.
Three F4 transgenic rice lines (L8-17, L12-13 and L7-15) were randomly selected for further study.
PCR analysis showed that 13 nitrogen-fixing genes could be inherited stably in L8-17 and L12-13 strains, while 13 nitrogen-fixing genes were separated 3:1 in L7-15 strains.
The results of rice genome resequencing showed that in L8-17 and L12-13 strains, 13 nitrogen-fixing genes were inserted into 2 loci of chromosome 1 of rice.
In L7-15, 13 nitrogen-fixing genes were inserted into two different chromosomes of rice.
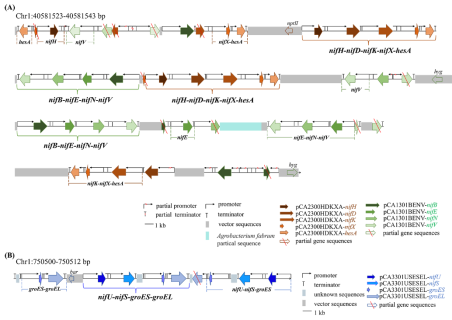
13 nitrogen fixing genes (nif) in F4 transgenic rice line L8-17 were inserted at 2 insertion sites of rice chromosome 1 and their arrangement.
(A) Insertion sites and arrangement of nine nitrogen fixing genes (nifH nifD nifK nifX hesA nifB nifE nifN nifV) on chromosome 1 of rice.
(B) Insertion sites and arrangement of four nitrogen fixing genes (nifU nifS groES groEL) on rice chromosome 1.
qRT-PCR analysis showed that 13 nitrogen-fixing genes were transcribed at different levels in three F4 transgenic rice lines (L8-17, L12-13 and L7-15).
Western blot analysis showed that 11 Nif proteins (NifB, NifH, NifD, NifK, NifE, NifN, NifV, NifX, HesA, NifS and GroEL) were stably expressed in the cytoplasm of rice line L8-17.
Moreover, NifD and NifK can form stable NifDK tetramer in rice cytoplasm, which is necessary for the formation of active nitrogenase.
At the same time, it was found that the molecular weight of NifH protein synthesized in rice cytoplasm was about 1.8 kDa smaller than that of P. polymyxa NifH, suggesting that NifH was cut by proteases in plant cytoplasm.
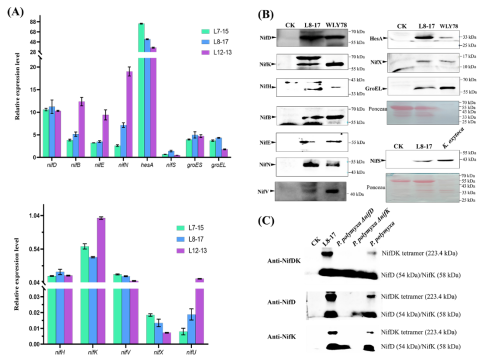
Transcription of nitrogen fixing gene (nif) and expression of nitrogenase protein (Nif) in F4 transgenic rice.
(A) qRT-PCR analysis of 13 nitrogen-fixing genes (nifB nifH nifD nifK nifE nifN nifX hesA nifV nifS nifU groES groEL) in 3 F4 transgenic rice lines L8-17 and L12-13 Transcription levels in L7-15.
(B) Western blot analysis of the expression of 11 Nif proteins (NifB, NifH, NifD, NifK, NifE, NifN, NifV, HesA, NifX, NifS and GroEL) in F4 transgenic rice line L8-17.
(C) Western blot (anaerobic and non-denaturing electrophoresis) analysis of the synthesis of NifDK tetrimers in F4 transgenic rice line L8-17.
In this study, the molecular weight of NifH protein expressed in rice cytoplasm decreased.
In order to solve the problem of NifH instability in plant cytoplasm, the amino acid sequence of NifH protein was studied with serial deletion and amino acid substitution, and the transient expression system of tobacco smoke was used. The results showed that the cleavage of NifH occurred between the 17th threonine (T) and the 18th filament amino acid (S).
Thirteen nifH amino acid substitution mutants (T17A, T17C, T17I, T17M, T17S, T17V, S18A, S18C, S18G, S18L, S18Q, S18T and Q19A) could restore the nitrogenase activity of P. Polymyxa_δNIFH mutant. In particular, three NifH mutants (T17C, T17V and S18A) could restore ΔnifH mutants to 59.4-71.9% nitrogenase activity, respectively.
Transient expression in tobacco also demonstrated that the 13 NifH amino acid replacement mutants were no longer cleaved by proteases in tobacco cytoplasm.
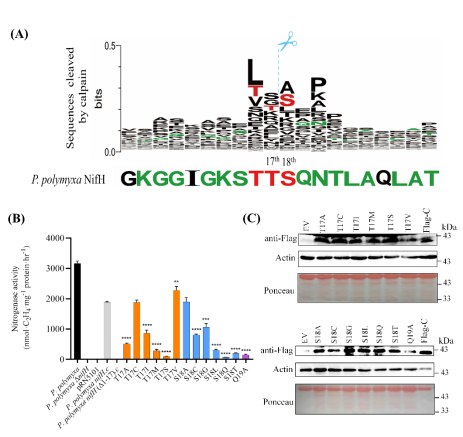
The cleavage site of NifH protein in plant cells and the NifH amino acid replacement mutant resistant to the cleavage of plant protease.
(A) Cleavage sites of NifH protein in plant cells.
(B) Nitrogenase activity of 13 nifH amino acid replacement mutants complementary to P. Polymyxa_δNifh mutants.
(C) 13 NifH amino acid substitution mutants were no longer cleaved by proteases in tobacco cytoplasm.
This study is the first time that the synthesis pathway of nitrogenase composed of 13 genes has been introduced into rice, which provides an important reference for realizing "autonomous nitrogen fixation" in gramineous plants.
At the same time, it provides an effective way to predict and solve the instability of nitrogenase components and other prokaryotic proteins in plant cytoplasm.