Design principles of strongly inducible synthetic promoters in yeast
DNA is the blueprint of life and controls what cells produce.
DNA also contains switches that tell these cells when to produce something and how much to produce.
Therefore, when introducing new genes into cells to produce useful chemicals, it is also necessary to include a genetic switch, a section of DNA called a "promoter."
In the field of synthetic biology, inducible promoters are often of irreplaceable importance because they can precisely control gene expression.
However, due to the high level of mechanistic complexity of eukaryotic promoters, designing eukaryotic promoters is often more challenging compared to prokaryotic promoters.
Leakage of inducible synthetic promoters (iSynP) in yeast is often caused by transcriptional readthrough of upstream random promoters and long-distance activation of iSynP by endogenous transcriptional activators from sequences at 1 kbp upstream of iSynP.

Without proper isolation, an iSynP designed from scratch could suffer a serious leak due to one or both of these events.
At present, the optimal design of iSynP remains unclear, and even with newly developed model-driven optimization techniques, designed promoters rarely outperform benchmark promoters in terms of inducibility.
Recently, Jun Ishii's team from Kobe University in Japan published a research paper entitled "Designing strong inducible synthetic promoters in yeasts" in Nature Communications.
They propose three design principles for yeast promoters:
Insert > 1 kbp insulator sequence to prevent transcriptional activation of upstream cryptic activation sequence;
Directly fuse operons upstream of TATA box;
Adding operon duplicates and/or screening (mutated) bacterial operons to reduce their covert activation without compromising binding to synthetic transcriptional activators (STAs) provides flexible guidelines for effective control of microbial production.
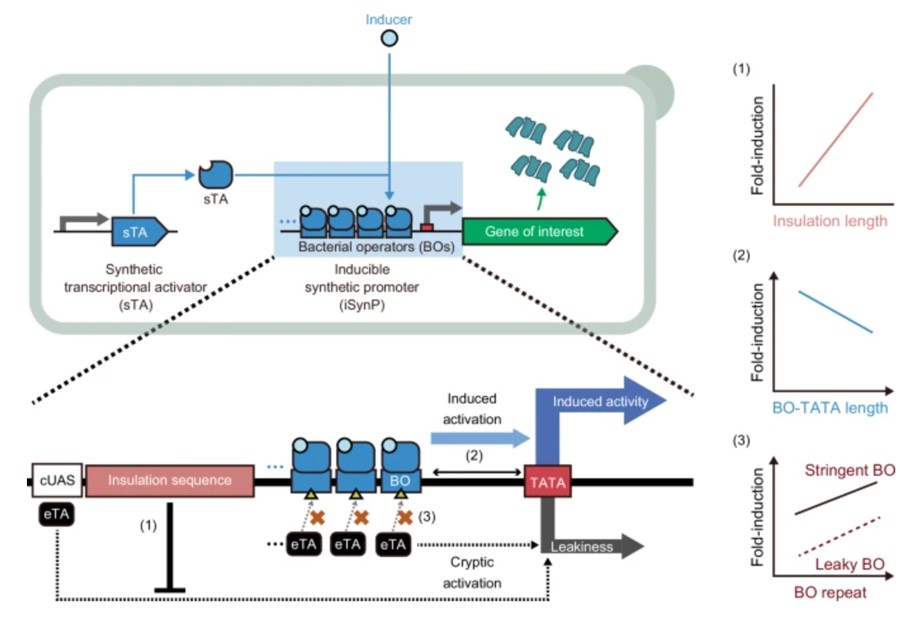
In order to design a strong iSynP suitable for yeast with minimal leakage, the researchers constructed DAPG-iSynP as a model promoter for methyl-trophic yeast Komagataella phaffii and found that the model was highly expressed in the absence of inducers.
The researchers speculate that remote transcriptional activation is the main cause of leakage.
So they tried to reduce leakage by inserting long insulation sequences.
For example, by inserting K.HaffII DNA fragments of different lengths upstream of iSynP, leakage was significantly reduced by up to 376-fold in a length-dependent manner, but induction activity was only slightly reduced (< 1.6-fold) when the length of the inserted fragment was up to 1.6 kb.
The researchers then decided to build a strongly induced iSynP and optimize the iSynP architecture through a series of strategies.
By carefully adjusting the length of the interval between the phlO and the TATA box and the start codon, the researchers found that a specific interval length can effectively increase the induction multiple.
For example, 94bp iSynP constructed in K. phaffii contains 53bp KpAOX1 core promoter. In the presence of DAPG, its induction multiple is as high as 1731±60 times, which is significantly stronger than that of conventional KpGAPDH promoter. Moreover, this method is universal. In Saccharomyces cerevisiae, the researchers also successfully used this method to construct an effective yeast iSynP.
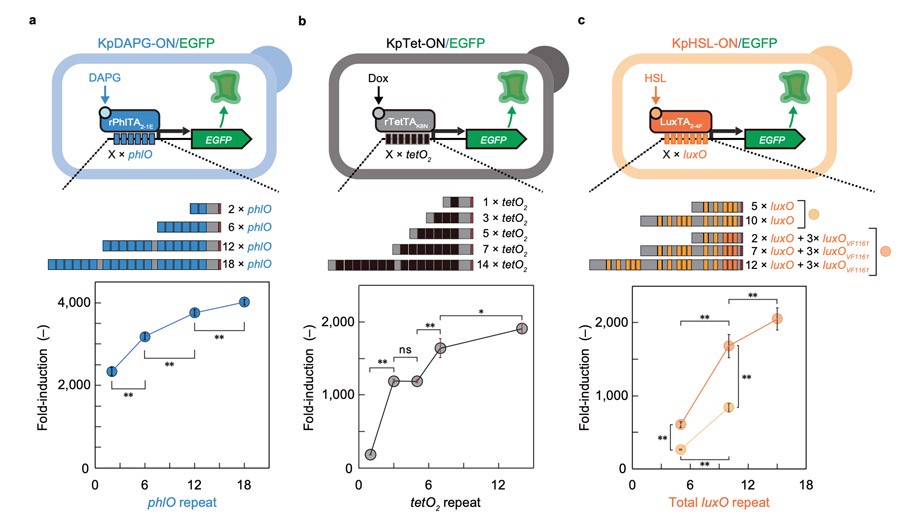
The researchers then constructed a variety of inducible promoters using different synthetic transcriptional activators (STAs) and corresponding bacterial operators.
They found that increasing the number of operon repeats not only increased the induction multiplier, but also reduced leakage.
Taking KpDAPG-ON switch as an example, when DAPG is added and two or more phlO repeats are merged with TATA box, the induction multiple of GFP expression can reach >2000 times, and the leakage is negligible.
KpTet-ON switches exhibit > 1000-fold induction of GFP expression when 3 or more tetO₂ repeats fuse with TATA-box.
To validate the application potential of this newly constructed strongly inducible iSynPs, the KpDAPG-ON system was used to produce a variety of drug proteins, including nanoantibodies ® ALX-0171 (gontivimab), an effective drug for the treatment of respiratory syncytial virus infection;
Nanoantibody ® ALX-0081 (also known as caplacizumab or Cablivi®) is used for the treatment of acquired thrombotic thrombocytopenic purpura.
The researchers were able to produce the two medicinal proteins not only in different yeast strains, but also in the same strain, and were able to independently control which biologics to produce at any time.
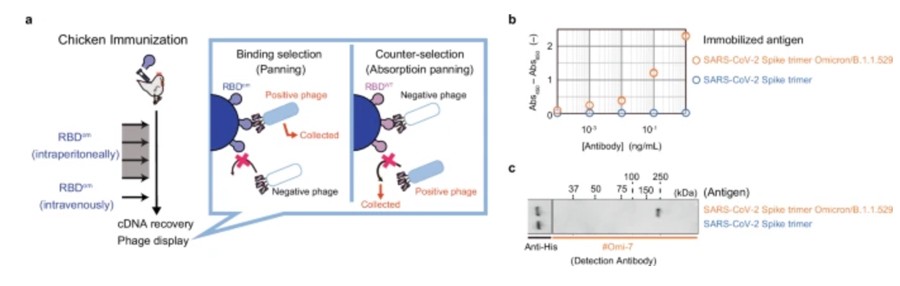
In addition, the researchers achieved the production of SARS-CoV-2 vaccine candidate proteins at a yield sufficient for chicken immunization to produce antibodies without even having to optimize fermentation and purification steps.
This fully proves that the system has great application potential in biomedicine.
In summary, through in-depth systematic analysis and optimization, the research team identified the key factors affecting the performance of yeast iSynPs, and established an effective strategy for constructing strongly induced iSynPs.
In the future, research can be carried out to further simplify the design of iSynP, improve its performance, and expand its application in multi-gene pathways.
References: 1、https://www.nature.com/articles/s41467-024-54865-z